Scientists have painted a bull’s-eye on a new target in the AIDS virus, a triumph not only for authors of a paper in the current issue of Nature but also for NAMD, a molecular modeling software package.
In a cover-featured Nature letter (May 30, 2013), the researchers say they’ve created an atom-by-atom computer model of a vital part of HIV, making its structure available to developers of antiviral drugs.
During the past two decades, the Department of Energy has supported NAMD through its INCITE Program, leadership computing facilities and Computational Science Graduate Fellowship (DOE CSGF).
“DOE machines have been the primary development platform for NAMD’s petascale simulation capabilities over the years,” says James Phillips, the code’s lead developer and a senior research programmer at the University of Illinois’ Beckman Institute. Major NAMD capabilities, he says, were developed on Jaguar, a Cray XT at Oak Ridge National Laboratory’s Leadership Computing Facility, and Mira, an IBM Blue Gene/Q at Argonne National Laboratory’s Leadership Computing Facility.
The Nature study was funded by the National Institutes of Health’s National Institute of General Medical Sciences and the National Science Foundation.
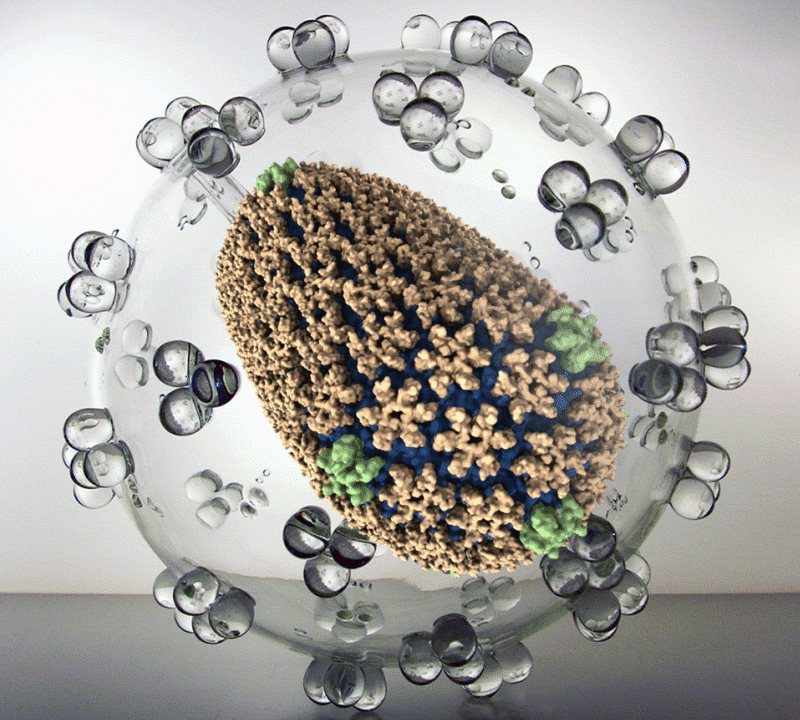
The new simulation reveals the structure of the HIV capsid – the delivery capsule the virus uses to insert its DNA into a host cell. Using the simulation, drug developers can study its construction, seeking places drugs could attack.
‘NAMD allows us to combine these two kinds of data so that we can resolve the whole structure in atomic detail.’
Team leader Klaus Schulten, Swanlund professor of physics at the University of Illinois at Urbana-Champaign, says the HIV capsid is “complicated machinery, and it’s a perfect target for fighting the virus. Now we can give this capsid’s fully molecularly characterized structure to the pharmacology community.”
The model is the latest of Schulten’s contributions using NAMD in biomedicine. In 2012, he and Laxmikant “Sanjay” Kale, professor of computer science at Illinois, shared the Sidney Fernbach Award from the Computer Society of the Institute of Electrical and Electronics Engineers. The honor recognized their work using NAMD and the parallel processing system Charm++ in biomolecular simulations.
A model of the human immunodefficiency virus type 1 (HIV-1) capsid is overlayed on an electron microscope image of the capsid. The capsid’s protein shell is believed to play a pivotal role in HIV infections, making it a prime target for drug therapies. Image, created with the VMD visualization package, courtesy of the Theoretical and Computational Biophysics Group, Beckman Institute, University of Illinois at Urbana-Champaign.
The HIV capsid may become a prime target in the search for new AIDS drugs. Other parts of the virus are under constant attack from a variety of antiviral medications. These targets continually evolve new means of drug resistance, but the capsid has evaded even an initial assault from modern medicine. It’s seen as an especially promising target because it’s apparently the weak spot attacked by immune systems in primates that defy HIV infection. But until now, no one had a means to tell which parts of it might be vulnerable.
The HIV capsid model was made possible through a long-term collaboration among the developers of a new Cray supercomputer named Blue Waters, Schulten’s team and NAMD’s developers. These researchers ensured that the software, hardware, experimental methods, and people progressed together to do just the kind of work the recent discovery embodies.
“We were approached to do this project because we were developing these capabilities for many years,” Phillips says.
NAMD’s creation began at Illinois in the mid-1990s, under DOE CSGF recipient William Humphrey. The idea was to write code for use on a variety of computers to model the structures of large, complex molecules important to biology and medicine.
A few years later, Phillips began work on the project, also as a doctoral student in the same fellowship. Phillips continued working on the project after earning his degree.
“I was encouraged to apply for the CSGF based on our group’s familiarity with the program through Bill Humphrey,” Phillips says. “I started the fellowship my third year, in 1995, and held it until I became a full-time lead NAMD developer in 1999.”
He recalls the valuable experiences he gained through coursework, scientific meetings and a summer research practicum at Los Alamos National Laboratory. “The total CSGF experience had an incredible impact on my development as a computational scientist,” he says.
Over the years, 37 developers have helped to expand and improve NAMD, Phillips says. More than 50,000 users worldwide have downloaded the software for free to run on machines ranging from PCs to supercomputers. Each time an update is released, 8,000 users download it.
Blue Waters offered an opportunity for NAMD to go beyond any modeling it had done before. The supercomputer was installed at the National Center for Supercomputing Applications at Illinois with a $208 million National Science Foundation grant.
“While Blue Waters only recently entered production, we have been using it for over a year for several biomolecular simulations,” Phillips says.
The HIV capsid model was one of these simulations and shows how NAMD enables researchers to exploit a supercomputer’s power.
The capsid is comprised of 216 six-sided protein structures and 12 five-sided ones and is shaped like a lopsided pillow. The model depicts about 64 million atoms.
During the simulation, Blue Waters and NAMD melded utterly different kinds of data. Some research techniques, such as X-ray crystallography, had shown the precise arrangements of atoms in the six-sided and five-sided protein structures. Other techniques, such as cryo-electron-microscopy, show the object on a larger scale. “What you get is basically a very fuzzy representation of the structure,” Phillips says.
Phillips compared the six- and five-sided structures to LEGO bricks and the overall structure to a castle made of the bricks. “You can see what the LEGO bricks look like, and you can see what the castle looks like,” he says, but you can’t see how the bricks fit together to form the castle. By combining the various kinds of data, the code created the complete picture. “NAMD allows us to combine these two kinds of data so that we can resolve the whole structure in atomic detail.”
Before its application to HIV, the code already proved invaluable for calculating numerous smaller molecular structures. Researchers at the Argonne Leadership Computing Facility (ALCF) have used NAMD to probe the secrets of important proteins.
For example, the modeling code has been used to simulate a variety of proteins that enable cells to communicate with one another and transport materials across cell membranes. These submicroscopic gates, hatches and revolving doors are vital in countless biological processes, from transmitting signals among brain cells to a heart’s beating to childbirth’s contractions.
Proteins in cell membrane are just as important in disease as they are in healthy activity. In one case, Argonne researchers used NAMD to show how the bacterial protein NDM-1 enables deadly microorganisms worldwide to quickly develop antibiotic resistance. This work is leading to new approaches in drug development.
In a separate instance, investigators at the San Diego Supercomputer Center, collaborating with ALCF scientists, used NAMD to reveal how the protein alpha-synuclein, a culprit in Parkinson’s and Alzheimer’s diseases, worms its way into neurons and links up with others of its kind to form cell-crippling pores through the cell membrane.
“NAMD has been and is being used by a number of projects that use the ALCF resources, some of which have contributed enhancements to the NAMD code,” says Paul Messina, the facility’s director of science. “NAMD is a shining example of the leverage that community codes provide to computational scientists: They can use the capabilities already in place and contribute modules that broaden the spectrum of problems that can be tackled with the code, as well as optimizations that increase scalability.”