Imagine trying to find someone in a New York City telephone book if the names were listed randomly.
Scientific databases prove even more daunting, perhaps holding a quadrillion bytes of data – about 5,000 desktop hard drives – or more. As databases grow, quickly finding specific information would be unmanageable without some data organization that facilitates searching.
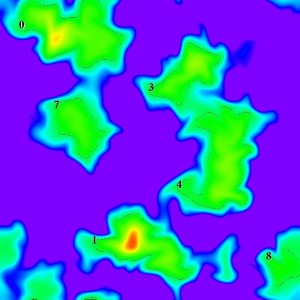
FastBit improves the capabilities of simulations such as this one looking at the chemical and physical features of combustion in time and space. A region-tracking algorithm helps researchers label and outline such things as areas of temperature differences and burning and non-burning over time. (Each panel represent regions of interest at selected intervals.)
Around the turn of the millennium, a team of scientists at Lawrence Berkeley National Laboratory (LBNL) began work on this database-indexing problem with something called FastBit. Since then, the technology has sped large data searches across the spectrum, from sifting through Enron e-mails and hunting new-drug molecules to tracking burn episodes in combustion simulations. The work earned the Berkeley researchers a 2008 R&D 100 Award for technology advances.
Many database indexes use tree data structures, such as B-trees. These look like upside-down trees, branching out into ever-more categories as the index goes deeper into a database. A search goes down the tree to find a specific section, sections within a section and so on. Such indexes work well when data are frequently added or deleted.
“B-trees are designed to support changes made to the data,” says Arie Shoshani, senior staff scientist at LBNL’s High Performance Computing Research Department in the Computational Research Division. “They make it possible to go down the tree and find the page or section where a change is made, and you only touch that section.”
But he adds, “The cost is that more space is needed for accommodating data changes. For scientific data that do not change over time, the extra space is wasteful and slows down the search.”
Bitmaps provide an alternative indexing approach. “We have been working on the idea of using bitmaps for scientific applications,” says Kesheng “John” Wu, who works in the Scientific Data Management Research Group at LBNL. “This method is known to work well for low-cardinality data, like the gender of a customer: male, female or unknown.”
For the most part, though, bitmaps only work efficiently when the cardinality is below, say, 100 distinct values for an attribute. “We wanted a way to make bitmaps work when the cardinality is higher, such as with instruments recording temperature or pressure,” Wu says.
That desire spawned FastBit.
A bitmap consists of a sequence of bits: 0s and 1s. Then, a set of bitmaps makes up a bitmap index. Typically, a bitmap index includes one bitmap for every value of an attribute. If you have a gender database of subjects, for example, it needs two bitmaps: one for female and one for male. It might look like this:
| Subject Gender | Female Bitmap | Male Bitmap |
|---|---|---|
| female | 1 | 0 |
| male | 0 | 1 |
| male | 0 | 1 |
| unspecified | 0 | 0 |
| female | 1 | 0 |
In combination, the female bitmap (10001) and the male bitmap (01100) make up the bitmap index that represents the data for these five subjects.
In scientific databases, an attribute – say, temperature – could easily require hundreds or thousands of distinct values. To encode temperatures from 0 to 100 as integers, for instance, requires 101 bitmaps. Many more values might be used if temperatures are computed as floating-point values in a simulation, and the range of measurements can be far greater than 0 to 100. These databases, therefore, could have bitmaps containing millions or billions of 0s and 1s.
Consequently, some approaches put the raw data in bins. In the temperature example, the bins might be for temperatures of 0-5 degrees, 5-10, 10-15 and so on, which would reduce the number of bitmaps to 20. Such binning, though, reduces the accuracy of later analysis if only the index is used.
Unlike tree structures, however, bitmap indexes do not work well with databases that change frequently. A modification to a record might lead to changes in a number of different bitmaps. When the bitmaps are compressed, as they usually are, these changes could require a significant amount of work. On the other hand, updating the bitmap indexes after appending a larger number of records is typically more efficient than updating tree-based indexes.
The example above also shows that bitmap indexes often contain many 0s, providing an avenue to significantly compress them. FastBit researchers found a way to specially compress the bitmaps, thus permitting more refined bins.
FastBit outruns other search indexes by a factor of 10 to 100 and doesn’t require much more room than the original data size.
A team of scientists – Wu, Shoshani and two LBNL scientists, Ekow Otoo and Kurt Stockinger – developed FastBit. To keep the size of FastBit manageable, it needed compressing. Wu and his colleagues tried a few approaches, including the byte-aligned bitmap code (BBC), which is used in Oracle.
But Wu and his colleagues saw a problem with using bytes. “Inside a computer, if you tell it to work with a byte, like with BBC, it pulls in an entire word and then extracts the byte. While byte alignment can be more compact, we thought we should use the word as the basic unit, and ensure that we do not break the word. That is the basic idea of word-aligned hybrid code, or WAH.”
It turns out that the feature of compressing the bitmaps without breaking the word boundaries greatly reduces the time needed to perform operations on the bitmaps, at a small cost of increasing the index size. That alone, however, wasn’t enough to achieve high search efficiency. The compression into words required a special selection of a word length.
Wu, Shoshani, and Otoo originally developed WAH to search data generated by the Department of Energy high-energy physics experiment called STAR. Instead of grabbing data by the byte, WAH organizes data by word – specifically, the native word length of the computer being used. WAH can grab one word – say, 32 bits, depending on the CPU – in less time than BBC uses to gather one byte. In fact, WAH can be configured to work with words that are 8, 16, 32, 64, 128 or more bits in length.
How does WAH work with 32-bit words? Rather than 32 bits, WAH compression uses groups of 31. WAH starts with a bitmap of thousands or millions of 0s and 1s and parses it into 31-bit groups. These groups could consist of all 0s, all 1s, or a combination. If a 31-bit group is a mixture of 0s and 1s, WAH stores the literal sequence, making a so-called literal word. Moreover, the most significant bit (MSB) is set to 0, which indicates that it’s a literal word, and the remaining 31 bits consist of the actual sequence from the bitmap. If a 31-bit sequence is all 0s or all 1s, then WAH creates a fill word. In such cases, WAH sets the MSB to 1, indicating a fill word. Then, the second MSB is the fill bit: 0 if the word consists of all 0s, and 1 if the word consists of all 1s. The remaining 30 bits encode the length of a fill. The length is an integer representing the number of 31-bit groups. A WAH-encoded fill could stretch across many words if a bitmap includes a long string of 0s or 1s. When WAH gets to the last word, which is generally only partially filled, it gets stored as a literal word.
By using 31-bit counts, the compressed data in fill words align exactly with uncompressed literal words (since one bit is used to distinguish between a literal and fill word), resulting in very efficient logical operations that can use the compressed fills directly, without decompressing them.
Wu explains: “With WAH, both compressed and decompressed indexes are word-aligned and generate results as compressed bitmaps that are word-aligned. That way, we avoid the need to look at individual bits in compressed data.” For example, to do a bitwise logical operation on two words that are fills, the software knows what every bit is, and there’s no need to decompress the data to run the operation. Likewise, literal words keep the bits available for easy bitwise logical operations.
How much WAH compresses a bitmap depends on the stretches of 1s or 0s in it. “On a typical scientific dataset, where the data are skewed,” Shoshani says, “the compressed data are about 30 percent of the size of the original. By comparison, B-trees tend to be two to four times larger than the original data.”
The upshot: FastBit outruns other search indexes by a factor of 10 to 100 and doesn’t require much more room than the original data size. With one million events from STAR, for example, Wu and his colleagues took the dozen most commonly queried attributes (from more than 500), built a bitmap for each and then stored the bit sequences in several ways, including BBC and WAH. On average, WAH searched the data 12 times faster than BBC.
The scientists received a patent for the WAH concept on Dec. 14, 2004.
FastBit already speeds up a wide range of searches. For example, the LBNL group tested FastBit on a portion of an Enron employee e-mail database made public during 2003 legal proceedings against the company. In January 2006, Wu, Shoshani and colleagues reported on a search-speed test on 250,000 Enron e-mails indexed with FastBit versus the open-source database management system MySQL. Depending on the specific parameters of a search, FastBit outran MySQL by 10 to 1,000 times.
“Most of the queries people use so far are on single sets of data,” Wu says. “The idea of exploring the Enron data was to work with more than one set, more than one table of data.” So to mine the entire collection of Enron data, tables had to be combined.
“Doing a join in databases is always a challenging problem,” Wu says. In addition, the bulk of Enron data is text instead of numbers. “A typical search on text data is the keyword search, and we wanted to explore that.”
FastBit also improves the capabilities of searching simulation data, such as looking at the chemical and physical features of combustion. “Here you are simulating what happened in space and time,” Wu says. For example, scientists might look for where the burning goes on and where it doesn’t and how that affects the byproducts generated in the process.
“You get lots of numbers, like temperature, but no one number that says, ‘I’m burning,’” Wu explains. “Even the highest temperature is not an indication of burning. The definition of burning involves a bunch of variables, and it’s complicated.” FastBit helps combustion scientists search through databases for the collection of parameters that do indicate burning and the quality of the burn.
Other applications push FastBit into medicine. For example, Jochen Schlosser, a doctoral student in bioinformatics at the University of Hamburg, Germany, uses FastBit to improve the discovery of new drugs. If a particular protein plays some role in a disease, for example, scientists search for a molecule that blocks said protein from doing its disease-related task. A large pharmaceutical company, though, might have a million or more compounds in a potential-drug library, and testing them all for a structural fit with every disease target takes immense amounts of computation.
So Schlosser and his colleagues developed TrixX BMI. This software generates information about every compound – so-called TrixX descriptors – that get stored in a compressed bitmap index. “You build an abstract of the compound and target protein to decide on an abstract level if the compound is a match or not,” Schlosser says. That abstract description encodes shape, molecular interactions and directional constraints in roughly 100 dimensions.
Instead of looking at every aspect of every compound in a library, TrixX BMI can eliminate compounds that have features that make them unlikely to dock with a specific protein. Using FastBit, the query phase of TrixX BMI runs 5 to 10 times faster than fully scanning the raw data.
“On average, the query phase is about one-fifth of the total runtime,” Schlosser says. Once FastBit finds compounds that look likely to dock with a specific protein, then Schlosser and his colleagues turn to further – and more computationally expensive – simulations.
FastBit’s capabilities make it useful for a wide range of problems. “Our primary goal at this point,” Wu says, “is trying to find new applications that could motivate our work. For example, we are currently working with some fusion-simulation groups, doing analysis of their data and dealing with interesting geometries.”
Likewise, some mechanical fine-tuning could lie ahead. FastBit grew out of research and it lacks some user amenities. “It was not exactly geared for production software,” Wu says. “If we get a chance, we might re-implement some of FastBit.”
Meanwhile, researchers keep putting FastBit to work, indexing and searching a wide range of high-dimensional and expansive databases. And the results will continue to teach us more about our world.